What is OMOP?
Last updated on 2026-02-08 | Edit this page
Overview
Questions
- What is OMOP?
- Why is using a standard important in healthcare data?
- How do OMOP tables relate to each other?
- What are concept_ids and how can we get an humanly readable name for them?
Objectives
- Examine the diagram of the OMOP tables and the data specification
- Understand OMOP standardization and vocabularies
- Connect to an OMOP database and explore the
concepttable - Get a humanly readable name for a concept_id
Setting up R
Getting started
The “Projects” interface in RStudio not only creates a working directory for you, but also remembers its location (allowing you to quickly navigate to it). The interface also (optionally) preserves custom settings and open files to make it easier to resume work after a break.
Connect to a database
For this episode we will be using the CDMConnector
package to connect to an OMOP Common Data Model database. We define a
function that will open this package and connect an appropriate dataset.
It is listed below but you will also find it in the
workshop/code/CDMConnector directory that you should have
downloaded. This package also contains synthetic example data that can
be used to demonstrate querying the data.
R
# Libraries
library(CDMConnector)
library(DBI)
library(duckdb)
library(dplyr)
library(dbplyr)
# Connect to GiBleed if not already connected
if (!exists("cdm") || !inherits(cdm, "cdm_reference")) {
db_name <- "GiBleed"
CDMConnector::requireEunomia(datasetName = db_name)
con <- DBI::dbConnect(duckdb::duckdb(),
dbdir = CDMConnector::eunomiaDir(datasetName = db_name))
cdm <- CDMConnector::cdmFromCon(con, cdmSchema = "main", writeSchema = "main")
}
OUTPUT
Download completed!Introduction
OMOP is a format for recording Electronic Healthcare Records. It allows you to follow a patient journey through a hospital by linking every aspect to a standard vocabulary thus enabling easy sharing of data between hospitals, trusts and even countries.
OMOP CDM Diagram
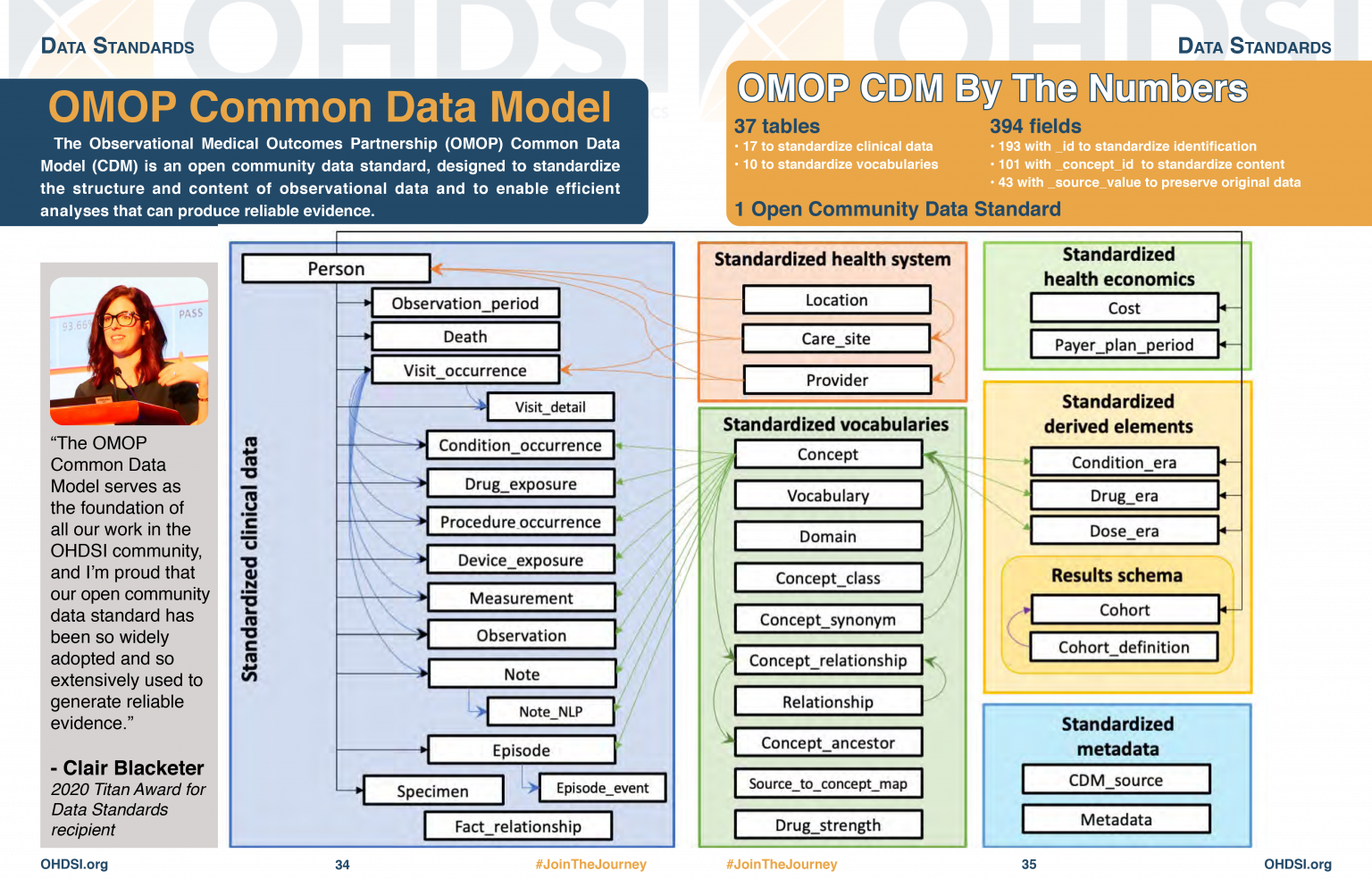
OMOP CDM stands for the Observational Medical Outcomes Partnership Common Data Model. You don’t really need to remember what OMOP stands for. Remembering that CDM stands for Common Data Model can help you remember that it is a data standard that can be applied to different data sources to create data in a Common (same) format. The table diagram will look confusing to start with but you can use data in the OMOP CDM without needing to understand (or populate) all 37 tables.
Challenge
Look at the OMOP-CDM figure and answer the following questions:
Which table is the key to all the other tables?
Which table allows you to distinguish between different stays in hospital?
The Person table
The Visit_occurrence table
Why use OMOP?
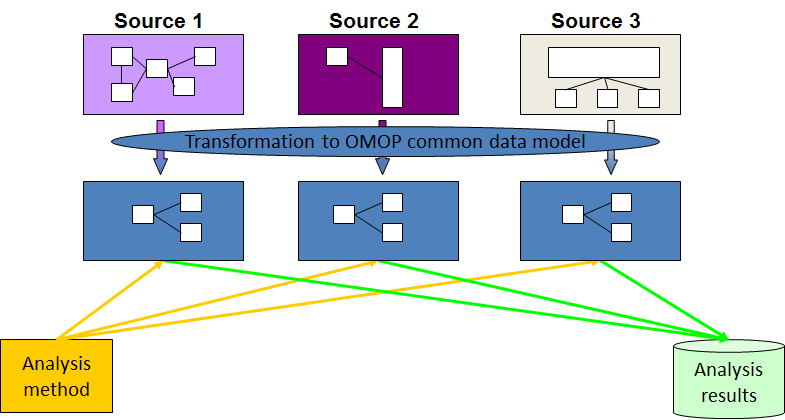
Once a database has been converted to the OMOP CDM, evidence can be generated using standardized analytics tools. This means that different tools can also be shared and reused. So using OMOP can help make your research FAIR.
Read in the database as above.
The data themselves are not actually read into the created cdm object. Rather it is a reference that allows us to query the data from the database.
Typing names(cdm) will give a summary of the tables in
the database and we can look at these individually using the
$ operator and the colnames command.
OMOP Tables
R
names(cdm)
OUTPUT
[1] "person" "observation_period" "visit_occurrence"
[4] "visit_detail" "condition_occurrence" "drug_exposure"
[7] "procedure_occurrence" "device_exposure" "measurement"
[10] "observation" "death" "note"
[13] "note_nlp" "specimen" "fact_relationship"
[16] "location" "care_site" "provider"
[19] "payer_plan_period" "cost" "drug_era"
[22] "dose_era" "condition_era" "metadata"
[25] "cdm_source" "concept" "vocabulary"
[28] "domain" "concept_class" "concept_relationship"
[31] "relationship" "concept_synonym" "concept_ancestor"
[34] "source_to_concept_map" "drug_strength" Looking at the column names in each table
R
colnames(cdm$person)
OUTPUT
[1] "person_id" "gender_concept_id"
[3] "year_of_birth" "month_of_birth"
[5] "day_of_birth" "birth_datetime"
[7] "race_concept_id" "ethnicity_concept_id"
[9] "location_id" "provider_id"
[11] "care_site_id" "person_source_value"
[13] "gender_source_value" "gender_source_concept_id"
[15] "race_source_value" "race_source_concept_id"
[17] "ethnicity_source_value" "ethnicity_source_concept_id"Challenge
How do you think the visit_occurrence table is used to
connect to the person table?
R
colnames(cdm$visit_occurrence)
OUTPUT
[1] "visit_occurrence_id" "person_id"
[3] "visit_concept_id" "visit_start_date"
[5] "visit_start_datetime" "visit_end_date"
[7] "visit_end_datetime" "visit_type_concept_id"
[9] "provider_id" "care_site_id"
[11] "visit_source_value" "visit_source_concept_id"
[13] "admitting_source_concept_id" "admitting_source_value"
[15] "discharge_to_concept_id" "discharge_to_source_value"
[17] "preceding_visit_occurrence_id"Looking at both tables we can see that they both have a column
labelled person_id which could be used to link them
together.
Notice that the visit_concept_id column in the
visit_occurrence table is also a concept_id. This
concept_id can be used to find out more information about the type of
visit (e.g. inpatient, outpatient etc) by looking it up in the
concept table. In this case the
visit_concept_id is 9201 which relates to an inpatient
visit. We can find this out by filtering the concept table
for concept_id 9201 and selecting the
concept_name column.
R
cdm$concept |>
filter(concept_id == 9201) |>
select(concept_name)
OUTPUT
# Source: SQL [?? x 1]
# Database: DuckDB 1.4.1 [unknown@Linux 6.8.0-1044-azure:R 4.5.2//tmp/RtmpY0FW18/file17431739c2fc.duckdb]
concept_name
<chr>
1 Inpatient VisitCODING_NOTE: We use filter to identify
the row we want and select to choose the column we want.
This is because we are querying a remote database, not one that is
local. If we were working with a local database we could just use
cdm$concept$concept_name[cdm$concept$concept_id == 9201] to
get the same result.
A useful function
Finding the humanly readable name for a concept_id will
be a useful function. We can create a function
get_concept_name() that takes a concept_id as
input and returns the concept_name.
Challenge
Create the function get_concept_name() that takes a
concept_id as input and returns the
concept_name.
R
get_concept_name <- function(id) {
cdm$concept |>
filter(concept_id == !!id) |>
select(concept_name) |>
pull()
}
Explanation of function code
- The function is called
get_concept_nameand it takes one argument,id. - Inside the function, we query the
concepttable from thecdmobject. - We use the
filterfunction to select rows where theconcept_idmatches the inputid. The!!operator is used to unquote the variable so that its value is used in the filter. - We then use
selectto choose only theconcept_namecolumn from the filtered results. - Finally, we use
pull()to extract theconcept_nameas a vector, which is returned by the function. We need to use this because we are querying a remote database, not one that is local.
Other useful tables
There are also other tables which will give you other information about concepts.
R
colnames(cdm$concept)
OUTPUT
[1] "concept_id" "concept_name" "domain_id" "vocabulary_id"
[5] "concept_class_id" "standard_concept" "concept_code" "valid_start_date"
[9] "valid_end_date" "invalid_reason" R
colnames(cdm$domain)
OUTPUT
[1] "domain_id" "domain_name" "domain_concept_id"R
colnames(cdm$vocabulary)
OUTPUT
[1] "vocabulary_id" "vocabulary_name" "vocabulary_reference"
[4] "vocabulary_version" "vocabulary_concept_id"- Using a standard makes it much easier to share data
- OMOP uses concepts to link different tables together
- The
concepttable contains humanly readable names for concept_ids
