Dates and times
Last updated on 2026-02-08 | Edit this page
Overview
Questions
- What
Objectives
- 1
Introduction
This episode considers dates and times in the OMOP Common Data Model (CDM).
For this episode we will be using a sample OMOP CDM database that is pre-loaded with data. This database is a simplified version of a real-world OMOP CDM database and is intended for educational purposes only.
(UCLH only) This will come in the same form as you would get data if you asked for a data extract via the SAFEHR platform (i.e. a set of parquet files).
As part of the setup prior to this course you were asked to download
and install the sample database. If you have not done this yet, please
refer to the setup instructions provided earlier in the course. For now,
we will assume that you have the sample OMOP CDM database available on
your local machine at the following path:
workshop/data/public/ and the functions in a folder
workshop/code.
You will then need to load the database as shown in the previous episode.
R
open_omop_dataset <- function(dir) {
open_omop_schema <- function(path) {
# iterate table level folders
list.dirs(path, recursive = FALSE) |>
# exclude folder name from path
# and use it as index for named list
purrr::set_names(~ basename(.)) |>
# "lazy-open" list of parquet files
# from specified folder
purrr::map(arrow::open_dataset)
}
# iterate top-level folders
list.dirs(dir, recursive = FALSE) |>
# exclude folder name from path
# and use it as index for named list
purrr::set_names(~ basename(.)) |>
purrr::map(open_omop_schema)
}
R
omop <- open_omop_dataset("./data/")
and the useful functions we created in the previous episode to look up concept names/ids.
R
library(arrow)
library(dplyr)
get_concept_name <- function(id) {
omop$public$concept |>
filter(concept_id == !!id) |>
select(concept_name) |>
collect()
}
R
get_concept_id <- function(name) {
omop$public$concept |>
filter(concept_name == !!name) |>
select(concept_id) |>
collect()
}
Dates and times in OMOP
Dates and times are used in most tables to record when events
happened,usually where they start and end date recorded. In many places
there is also a time component, for example to record the time of day
when a measurement was taken. Column names are frequently either
suffixed with _date or _datetime to indicate
whether they contain just a date or both date and time information.
However in our dataset dates and times are recorded as strings rather
than as date or datetime objects. This is because the parquet files
don’t have a standard way to store date and datetime objects, so they
are stored as strings in the format “YYYY-MM-DD” for dates and
“YYYY-MM-DD HH:MM:SS” for datetimes. When we read the data into R, we
can convert these strings to date or datetime objects using the
as.Date() or as.POSIXct() functions,
respectively. This will allow us to perform date and time calculations
and visualizations more easily.
Usually the dates come in pairs, for example
condition_start_date and condition_end_date in
the condition_occurrence table. This allows us to determine
the duration of a condition, for example, by calculating the difference
between the start and end dates.
Challenge
Using the condition_occurrence table, find the average
duration of conditions in this dataset.
R
# First we need to read in the condition_occurrence table
condition_occurrence <- omop$public$condition_occurrence |>
collect()
# Then we need to exclude any rows where the end date is missing, as we can't calculate the duration for these
condition_occurrence <- condition_occurrence |>
filter(!is.na(condition_end_date))
# Now we can calculate the duration of each condition by taking the difference between the end date and the start date
# TWO DO:fix this
condition_occurrence <- condition_occurrence |>
mutate(condition_duration = as.numeric(as.Date.numeric(condition_end_date) - as.Date.numeric(condition_start_date)))
ERROR
Error in `mutate()`:
ℹ In argument: `condition_duration =
as.numeric(as.Date.numeric(condition_end_date) -
as.Date.numeric(condition_start_date))`.
Caused by error in `unclass(x) * 86400`:
! non-numeric argument to binary operatorR
average_duration <- mean(condition_occurrence$condition_duration, na.rm = TRUE)
average_duration
OUTPUT
[1] NAIt is not uncommon for the end date to be missing in the data, for example if a condition is ongoing at the time of data extraction. In this case, we can only calculate the duration for conditions that have an end date recorded.
Also consider the measurement table where we have both a
measurement_date and a measurement_datetime
column. The measurement_date column contains just the date
of the measurement, while the measurement_datetime column
contains both the date and time of the measurement. Depending on the
analysis we want to do, we may choose to use one or the other of these
columns. Note generally a measurement doesn’t have a start and end date,
but just a single date or datetime when the measurement was taken.
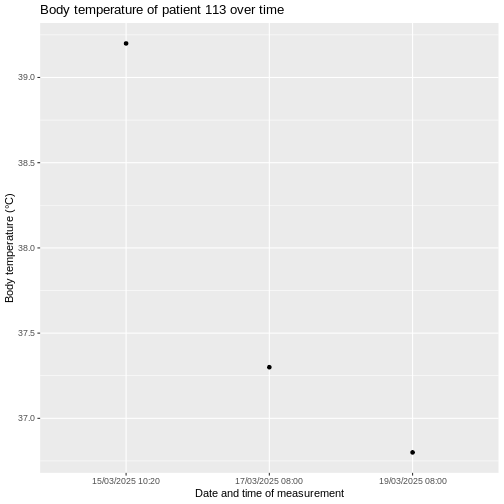
Challenge
Using the measurement table, graph the
Body temperature of patient 113over time. You
can use either the measurement_date or
measurement_datetime column for the x-axis.
R
library(ggplot2)
# First we need to know the concept id for body temperature
get_concept_id("Body temperature")
OUTPUT
# A tibble: 1 × 1
concept_id
<int>
1 3020891R
# Then we need to read in the measurement table, filter for the measurements of interest and collect the data into memory
measurement <- omop$public$measurement |>
filter(person_id == 1113 & measurement_concept_id == 3020891) |>
collect()
# Now we can plot the body temperature over time
ggplot(measurement, aes(x = measurement_datetime, y = value_as_number)) +
geom_line() +
geom_point() +
labs(title = "Body temperature of patient 113 over time",
x = "Date and time of measurement",
y = "Body temperature (°C)")
Dates and times are used in most tables in the OMOP CDM to record when events happened.
They are often used in pairs to record the start and end of an event, which allows us to calculate the duration of that event.
