Medications
Last updated on 2026-02-08 | Edit this page
Estimated time: 0 minutes
Overview
Questions
Where are medications stored ?
How do you trace the relationship between concepts from different vocabularies?
Objectives
Know that exposure of a patient to medications is mainly stored in the drug_exposure table
Understand that drug concepts can be at different levels of granularity
Understand that source values are mapped to a standard vocabulary
Introduction
This episode considers medications (the drug exposure table) in the OMOP Common Data Model (CDM).
For this episode we will be using a sample OMOP CDM database that is pre-loaded with data. This database is a simplified version of a real-world OMOP CDM database and is intended for educational purposes only.
(UCLH only) This will come in the same form as you would get data if you asked for a data extract via the SAFEHR platform (i.e. a set of parquet files).
As part of the setup prior to this course you were asked to download
and install the sample database. If you have not done this yet, please
refer to the setup instructions provided earlier in the course. For now,
we will assume that you have the sample OMOP CDM database available on
your local machine at the following path:
workshop/data/public/ and the functions in a folder
workshop/code.
You will then need to load the database as shown in the previous episode.
R
open_omop_dataset <- function(dir) {
open_omop_schema <- function(path) {
# iterate table level folders
list.dirs(path, recursive = FALSE) |>
# exclude folder name from path
# and use it as index for named list
purrr::set_names(~ basename(.)) |>
# "lazy-open" list of parquet files
# from specified folder
purrr::map(arrow::open_dataset)
}
# iterate top-level folders
list.dirs(dir, recursive = FALSE) |>
# exclude folder name from path
# and use it as index for named list
purrr::set_names(~ basename(.)) |>
purrr::map(open_omop_schema)
}
R
omop <- open_omop_dataset("./data/")
and the useful functions we created in the previous episode to look up concept names/ids.
R
library(arrow)
library(dplyr)
get_concept_name <- function(id) {
omop$public$concept |>
filter(concept_id == !!id) |>
select(concept_name) |>
collect()
}
R
get_concept_id <- function(name) {
omop$public$concept |>
filter(concept_name == !!name) |>
select(concept_id) |>
collect()
}
The OMOP drug_exposure
table stores exposure of a patient to medications. The purpose of
records in this table is to indicate an exposure to a certain drug as
best as possible. In this context a drug is defined as an active
ingredient. Drug Exposures are defined by Concepts from the Drug domain,
which form a complex hierarchy. As a result, one
drug_source_concept_id may map to multiple standard
concept_ids if it is a combination product. Records in this
table represent prescriptions written, prescriptions dispensed, and
drugs administered by a provider to name a few. The
drug_type_concept_id can be used to find and filter on
these types. This table includes additional information about the drug
products, the quantity given, and route of administration.
The main columns are :
| column name | content |
|---|---|
| drug_exposure_id | unique identifier given that person can get multiple exposures per visit |
| person_id | the patient |
| drug_concept_id | standard drug identifier, can be at different levels of granularity |
| drug_exposure_start_date | may also be an optional start_datetime |
| drug_exposure_end_date | may also be an optional end_datetime |
| drug_type_concept_id | where the record came from e.g. EHR administration record |
| quantity | the amount of drug given |
| route_concept_id | the route of administration e.g. oral, intravenous etc |
| visit_occurrence_id | the visit during which the drug was given |
| drug_source_concept_id | OMOP concept ID for the source value |
Drug data can be very complicated, as can the process of converting from the source data to OMOP. You may not find what you expect depending on this and the quality of the source data.
Drug concepts
The standard OHDSI drug vocabularies are called RxNorm
and RxNormExtension. RxNorm contains all drugs
currently on the US market. RxNormExtension is maintained
by the OHDSI community and contains all other drugs.
A particular concept_id can be at one of a number of different levels in a drug hierarchy.
Challenge
List the main levels of drug concepts in RxNorm.
R
omop$public$concept |>
filter(vocabulary_id == "RxNorm") |>
select(concept_class_id) |>
collect() |>
distinct() |>
arrange(concept_class_id)
OUTPUT
# A tibble: 4 × 1
concept_class_id
<chr>
1 Clinical Drug
2 Clinical Drug Form
3 Ingredient
4 Quant Branded DrugAnswer: There are more levels than shown here, but that is a disadvantage of using a small sample database. In a full OMOP CDM database you would see more levels.
A fuller example of the drug concept hierarchy in RxNorm is shown in the table below.
| RxNorm concept_class_id | Description |
|---|---|
| Clinical Drug | A combination of an ingredient, strength, and dose form (e.g., Ibuprofen 200 mg Oral Tablet). |
| Clinical Drug Comp | A drug component with strength but no form (e.g., Ibuprofen 200 mg). |
| Clinical Drug Form | A drug with a specific dose form but no strength (e.g., Ibuprofen Oral Tablet). |
| Quant Clinical Drug | A clinical drug with a specific quantity (e.g., Ibuprofen 200 mg Oral Tablet 1). |
| Ingredient | A base active drug ingredient, without strength or dose form (e.g., Ibuprofen). |
There are also concepts for Branded drugs and for packs of drugs (e.g. a box of 30 tablets) but these are not shown in this sample table.
Drug mapping in the NHS
Drugs in the NHS are standardised to the NHS Dictionary of Medicines
and Devices (dm+d). dm+d is included in OMOP so there are values of OMOP
concept_id for each dm+d. However because dm+d is not a standard
vocabulary in OMOP it is translate once more to get to a standard OMOP
concept id in RxNorm or RxNormExtension that
can be used in collaborative studies. If there is a drug_concept_id
value of 0 and there are source codes this can be because that drug
doesn’t map to a standard ID. Reminder that the source values are stored
in these columns.
Challenge
Look up the concept_id 871182 and find
the corresponding RxNorm concept_id.
R
library(dplyr)
# make a copy of the concept table
concepts <- omop$public$concept |> collect()
# look up the concept entry
dmd_concept_1 <- concepts |>
filter(concept_id == 871182) |>
select(concept_id, concept_name, domain_id, vocabulary_id, concept_class_id) |>
collect()
dmd_concept_1
OUTPUT
# A tibble: 1 × 5
concept_id concept_name domain_id vocabulary_id concept_class_id
<int> <chr> <chr> <chr> <chr>
1 871182 Lorazepam 1mg tablets Drug dm+d VMP R
# this is the dose of lorazepam
# now look up any concepts that have a similar name
similar <- filter(concepts, grepl('Lorazepam', concept_name, TRUE))
similar
OUTPUT
# A tibble: 4 × 6
concept_id concept_name domain_id vocabulary_id standard_concept
<int> <chr> <chr> <chr> <chr>
1 871182 Lorazepam 1mg tablets Drug dm+d ""
2 19019113 lorazepam MG Oral Tablet Drug RxNorm "S"
3 35777064 1 ML Lorazepam 4 MG/ML In… Drug RxNorm Exten… "S"
4 36816707 Lorazepam 4mg/1ml solutio… Drug dm+d ""
# ℹ 1 more variable: concept_class_id <chr>Answer: We can see from the resulting table
that there are entries for each lorazepam dose from both the dm+d and
RxNorm vocabularies. The concept_id 871182
corresponds to the dm+d concept “Lorazepam 1mg tablets”. This seems maps
to the RxNorm concept “lorazepam Oral Tablet” which has
concept_id 19019113, but without knowing
the quantity we can’t be sure which dose it maps to. This is an example
of the complexity of drug data and the mapping process.
CODING_NOTE: The function grepl() is
used to find all concepts that have “Lorazepam” in their name. We have
added the ignore.case = TRUE argument to make the search
case-insensitive. This allows us to find all relevant concepts
regardless of how they are capitalized in the concept names.
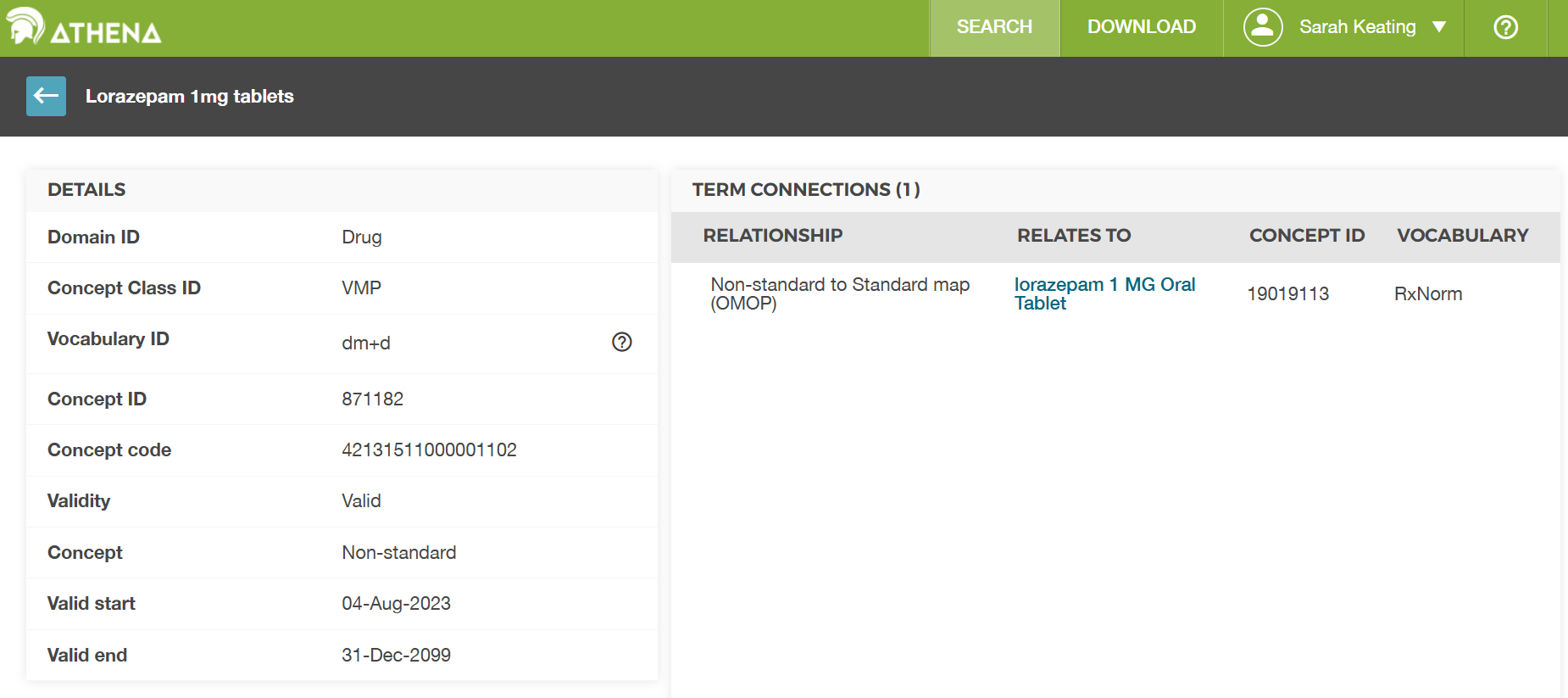
Looking at the entry for concept_id
871182 we can see that it is in the dm+d vocabulary. We
can see that is connected to the RxNorm concept “lorazepam 1 MG Oral
Tablet” which has concept_id 19019113. So
our assumption above was correct, but the concept table in our dataset
didn’t fill in the name fully so we couldn’t be sure without looking it
up in the official table.
TO TO: We need an example on quantity and route of administration to show how these are stored in the drug_exposure table and how they can be looked up in the concept table.
- Know that exposure of a patient to medications is mainly stored in the drug_exposure table
- Understand that drug concepts can be at different levels of granularity
- Understand that source values are mapped to a standard vocabulary
